Model:
Parameters: 1.7B
Architecture: Joint-Embedding Predictive Architecture (JEPA)
standardmodelbio/SMB-v1-1.7B-StructureParameters: 1.7B
Architecture: Joint-Embedding Predictive Architecture (JEPA)
Key Differentiators
State Prediction
Predicts future patient states in latent space, not text tokens
Causal Learning
Learns cause-and-effect: (Pre-State + Intervention) → Post-State
Multimodal Fusion
Unifies genomics, imaging, EHR, and proteomics
Environment Activation
Usage
Architecture
The Standard Model uses Joint-Embedding Predictive Architecture (JEPA) — treating the patient as a dynamic “world” and treatments as interventions that change that world.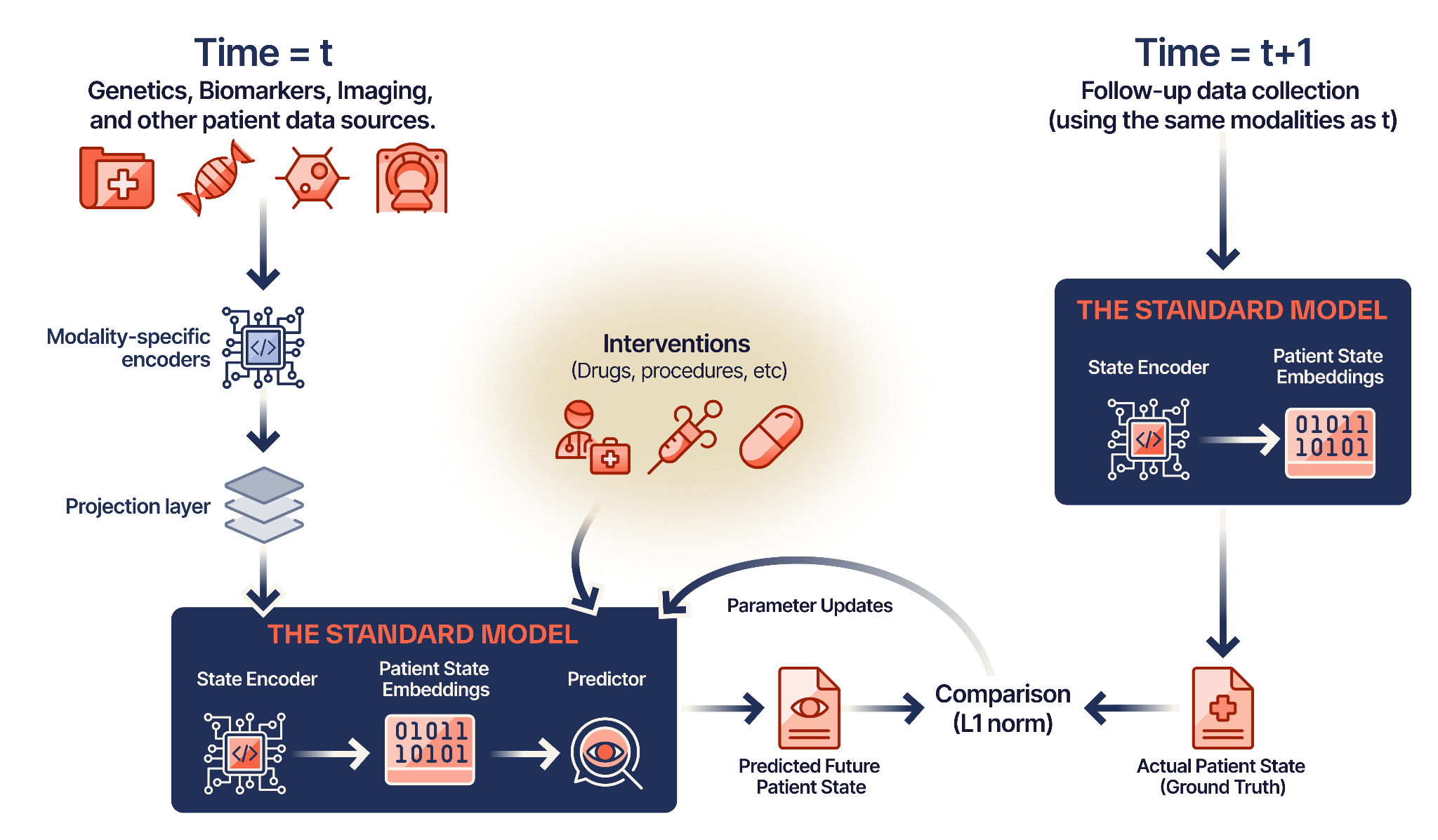
How It Works
1
Modality Ingestion
Raw signals — genomics, proteomics, imaging, EHR data — pass through modality-specific encoders. Each encoder is trained to extract meaningful representations from its data type.
2
Fusion Layer
A specialized projector maps these encodings into a universal latent space. This creates a “fused” patient state embedding that retains both high-level semantic context and low-level biological granularity.
3
State Prediction
Given the current patient state S(t) and an intervention A(t), the model predicts the future state S(t+1) in latent space — not as text, but as a dense embedding.
4
Hybrid Optimization
The model combines supervised fine-tuning (anchoring to clinical outcomes) with JEPA objectives (learning dynamics), preventing training collapse.
Extracting Embeddings
Get patient state embeddings for downstream tasks:Use Cases
Treatment Simulation
Simulate how a tumor would evolve under Treatment A versus Treatment B by conditioning on different interventions.
Trajectory Prediction
Predict disease progression over 3, 6, or 12 month windows.
Digital Twins
Create evolving patient representations that update as new data arrives.
Response Prediction
Model probability of response to specific therapies.
Memory Optimization
SMB-v1-Structure requires approximately 16GB GPU memory at full precision. Use these techniques to reduce memory:- Float16
- 8-bit Quantization
- 4-bit Quantization
Hardware Requirements
| Precision | GPU Memory | Recommended GPU |
|---|---|---|
| float32 | 16 GB | A100, A6000 |
| float16 | 8 GB | RTX 4090, A10 |
| 8-bit | 4 GB | RTX 3080, T4 |
| 4-bit | 2 GB | RTX 3060 |
Research
The Patient is Not a Document
Read the announcement blog post explaining the motivation and architecture behind SMB-v1-Structure.
