
New Release: SMB-v1-structure is now available — our flagship multimodal foundation model for oncology built on Joint-Embedding Predictive Architecture (JEPA).
The Problem with Current AI
For years, the approach to biomedical AI has been simple: take a massive general-purpose model, feed it medical text, and watch it pass licensing exams. But when tested on realistic patient cases requiring actual treatment decisions, GPT-4 achieves just 30.3% completeness. The disconnect lies in a single, flawed assumption: that language is a sufficient proxy for disease biology.Our Approach: A Biological World Model
The Standard Model represents a paradigm shift:| Traditional LLMs | Standard Model |
|---|---|
| Predicts tokens | Predicts patient states |
| Static snapshots | Dynamic trajectories |
| Text as proxy | Raw biological signals |
| Pattern matching | Causal reasoning |
From Description to Dynamics
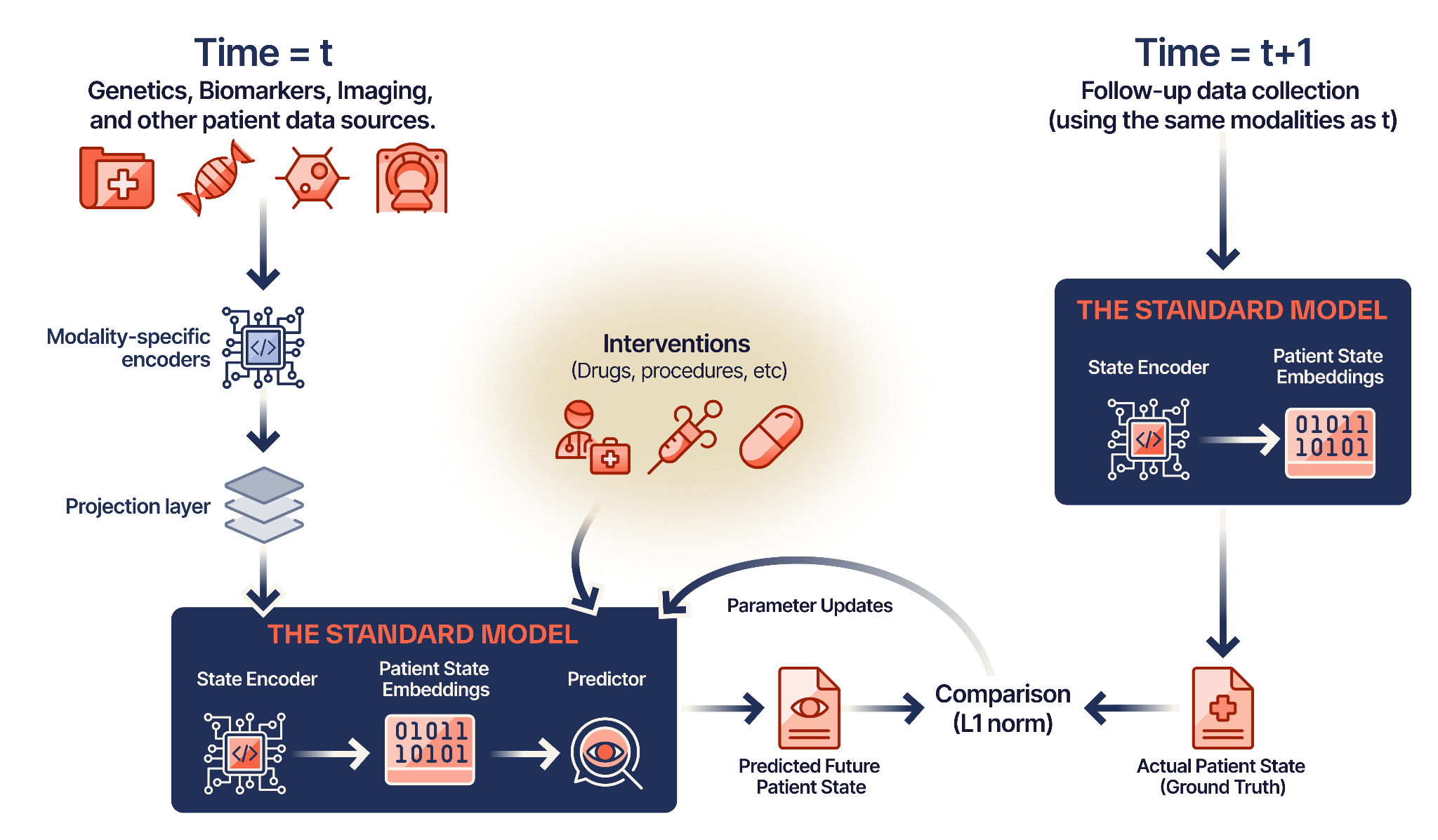
A clinician does not simply ask “Is this cancer?” (classification). Rather, they ask:“Given the patient’s current state S(t) and this specific intervention I, what will their state be in 6 months S(t+1)?”Our Standard Model answers this question by modeling biological dynamics, not generating text.
Architecture Overview
The Standard Model is built on Joint-Embedding Predictive Architecture (JEPA) — the same paradigm powering advances in robotics and autonomous driving, now applied to human biology.1
Modality Ingestion
Raw signals — genomics, proteomics, imaging, EHR data — pass through modality-specific encoders and fuse into a universal latent space.
2
State Prediction
Given the current patient state S(t) and an intervention A(t), the model predicts the future state S(t+1) in latent space.
3
Causal Learning
Training data is structured as (Pre-State + Intervention) → (Post-State), forcing the model to learn cause-and-effect relationships.
4
Hybrid Optimization
Supervised fine-tuning anchors the model to clinical outcomes while JEPA objectives learn the underlying dynamics.
Core Beliefs
Performance Is Everything
Performance Is Everything
Small differences in clinical efficacy relative to standard of care drive billions of dollars in revenue and change millions of lives. Every marginal improvement can define clinical, regulatory, or commercial success.
The Patient is Not a Document
The Patient is Not a Document
Biological signals exist at every scale — from molecular variations in genomic sequences to cellular structures in histopathology slides, to anatomical changes in CT volumes. Language alone cannot capture this complexity.
Dynamics Over Description
Dynamics Over Description
Clinical oncology is not a series of static snapshots; it is an evolving biological trajectory. We model how tumors progress and respond to therapy, not just what they look like at a single moment.
Multimodal by Nature
Multimodal by Nature
Patient data is inherently multimodal because patients are inherently multimodal. The best foundation models integrate data across modalities and disease areas.
Use Cases
Once the Standard Model learns a patient’s biological state, it can power diverse applications:Treatment Simulation
Simulate how a tumor would evolve under Treatment A versus Treatment B.
Digital Twins
Create evolving digital representations for prognosis and intervention planning.
Clinical Trial Optimization
Forecast trials in silico and optimize inclusion/exclusion criteria.
Progression Prediction
Predict disease trajectories and time-to-event outcomes.
Response Prediction
Model probability of response to specific therapies.
Toxicity Assessment
Predict treatment-related adverse events before they occur.
Model Families
SMB-v1
1.7B parameters · Flagship JEPA-based world model for oncology
SMB-EHR
Foundation models for electronic health records
SMB-Vision
Medical imaging encoders for radiology and pathology
SMB-Language
Biomedical language models for clinical text
Next Steps
Quickstart
Get up and running with Standard Model in minutes.
Models
Explore our full model catalog.
Research
Read our published papers and methodology.
Blog
Follow our latest research on Substack.
