Featured: SMB-v1-Structure
SMB-v1-Structure
1.7B parameters · Our flagship biological world modelThe first multimodal foundation model for oncology built on Joint-Embedding Predictive Architecture (JEPA). Predicts patient trajectories, not tokens.
What Makes It Different
State Prediction
Predicts future patient states in latent space, not text tokens.
Causal Learning
Learns cause-and-effect from (Pre-State + Intervention) → Post-State.
Multimodal Fusion
Ingests genomics, imaging, EHR, and proteomics into unified embeddings.
Architecture
The Standard Model uses a Joint-Embedding Predictive Architecture (JEPA) — treating the patient as a dynamic “world” and treatments as interventions that change that world.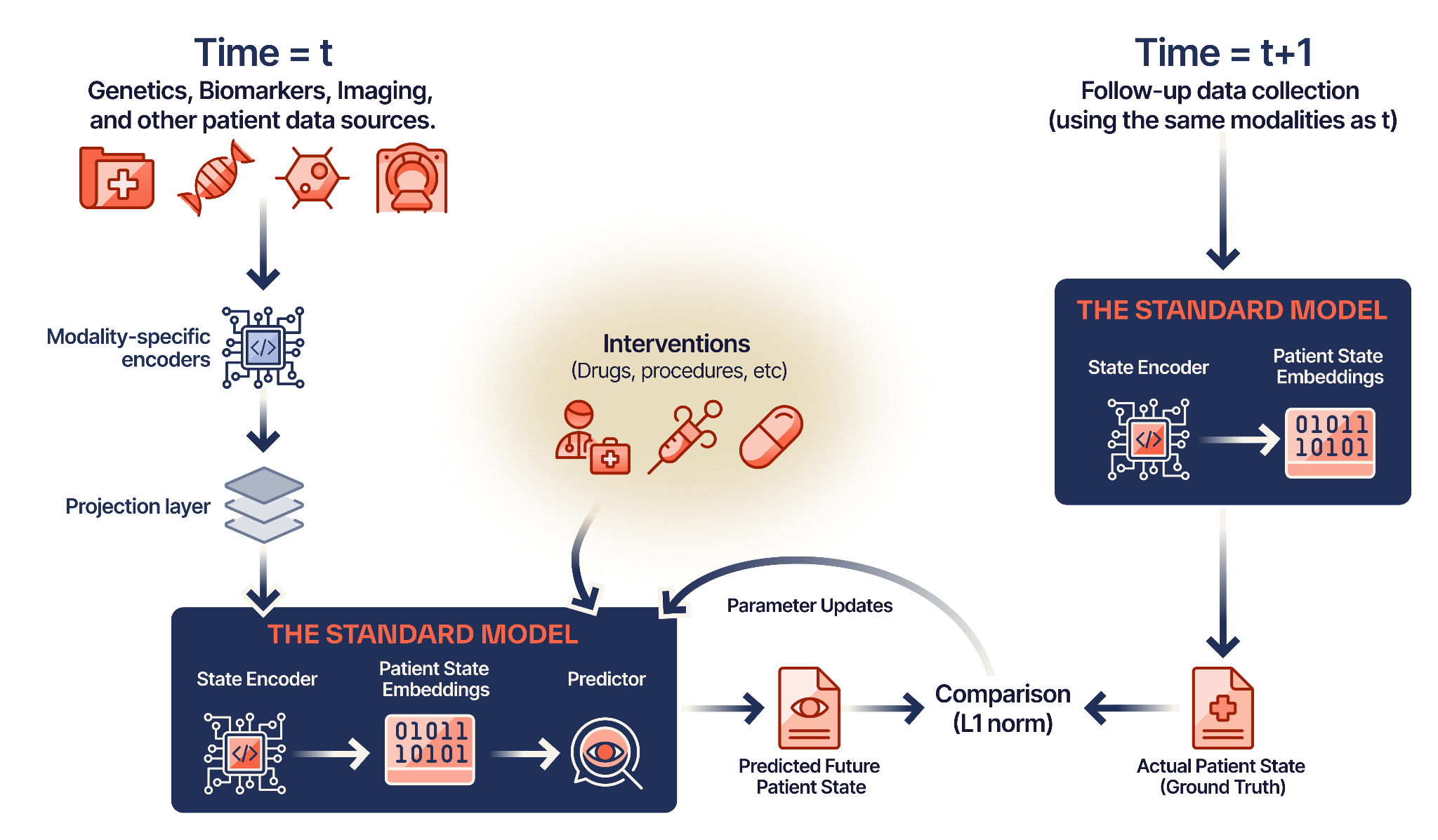
Model Families
SMB-EHR
Electronic health record foundation models for clinical event prediction.
SMB-Vision
Medical imaging foundation models for radiology and pathology.
SMB-Language
Biomedical language models for clinical text understanding.
SMB-v1
Flagship JEPA-based world models for oncology trajectories.
SMB-EHR
Foundation models for electronic health records, trained to predict clinical events and understand patient disease trajectories.smb-ehr-4b
4B parameters · EHR Foundation ModelReframes EHRs as timestamped chains of clinical events and predicts next events to improve temporal reasoning over disease trajectories.
SMB-Vision
Medical imaging foundation models trained on radiology and pathology data. These encoders power the vision capabilities of the Standard Model.smb-vision
97M - 600M parameters · Medical Imaging Foundation ModelsVision encoders for radiology, pathology, and CT imaging tasks. Multiple sizes available from base (97M) to large (600M) variants.
SMB-Language
Biomedical language models for clinical text understanding and sentence similarity.smb-mntp-llama-3.1-8b-v1
8B parameters · Sentence SimilarityFine-tuned Llama 3.1 for biomedical sentence similarity and text understanding.
Model Selection Guide
Patient Trajectory Prediction
Patient Trajectory Prediction
Use SMB-v1-Structure for predicting how patients will evolve over time, simulating treatment outcomes, and modeling disease dynamics.
Clinical Event Prediction
Clinical Event Prediction
Use smb-ehr-4b for next-event prediction, temporal reasoning over EHRs, and disease trajectory analysis from clinical records.
Medical Image Analysis
Medical Image Analysis
Start with smb-vision-base for general tasks. Use smb-vision-ct-base-0519 for CT-specific applications or smb-vision-v0-risk for risk stratification.
Clinical Text Understanding
Clinical Text Understanding
Use smb-mntp-llama-3.1-8b-v1 for semantic search, sentence similarity, and clinical text embeddings.
All Models
Complete catalog of available models:| Model | Family | Parameters | Task | Link |
|---|---|---|---|---|
| SMB-v1-1.7B-Structure | Standard Model | 1.7B | World Model / Trajectory Prediction | HuggingFace |
| smb-ehr-4b | SMB-EHR | 4B | EHR / Next Event Prediction | HuggingFace |
| smb-vision-v0-risk | SMB-Vision | 0.6B | Vision / Risk Assessment | HuggingFace |
| smb-vision-v0-mim | SMB-Vision | 0.6B | Vision / Masked Image Modeling | HuggingFace |
| smb-vision-large | SMB-Vision | 0.3B | Vision / General Encoder | HuggingFace |
| smb-vision-base | SMB-Vision | 97.2M | Vision / General Encoder | HuggingFace |
| smb-vision-ct-base-0519 | SMB-Vision | 97.2M | Vision / CT-Specific | HuggingFace |
| smb-vision-vjepa2-vitl-384-256 | SMB-Vision | 0.3B | Vision / V-JEPA2 | HuggingFace |
| smb-mntp-llama-3.1-8b-v1 | SMB-Language | 8B | Language / Sentence Similarity | HuggingFace |
Hardware Requirements
GPU memory requirements vary by model size. We recommend the following minimums:
| Model Size | Minimum GPU Memory | Recommended |
|---|---|---|
| ~100M (Base) | 4 GB | 8 GB |
| ~300M (Large) | 8 GB | 16 GB |
| ~600M (v0) | 12 GB | 24 GB |
| 1.7B (SMB-v1-Structure) | 16 GB | 32 GB |
| 4B (EHR) | 24 GB | 48 GB |
| 8B (Language) | 32 GB | 80 GB |
